
Identification of shared and unique mechanisms of atopic dermatitis and ulcerative colitis
What was the goal?
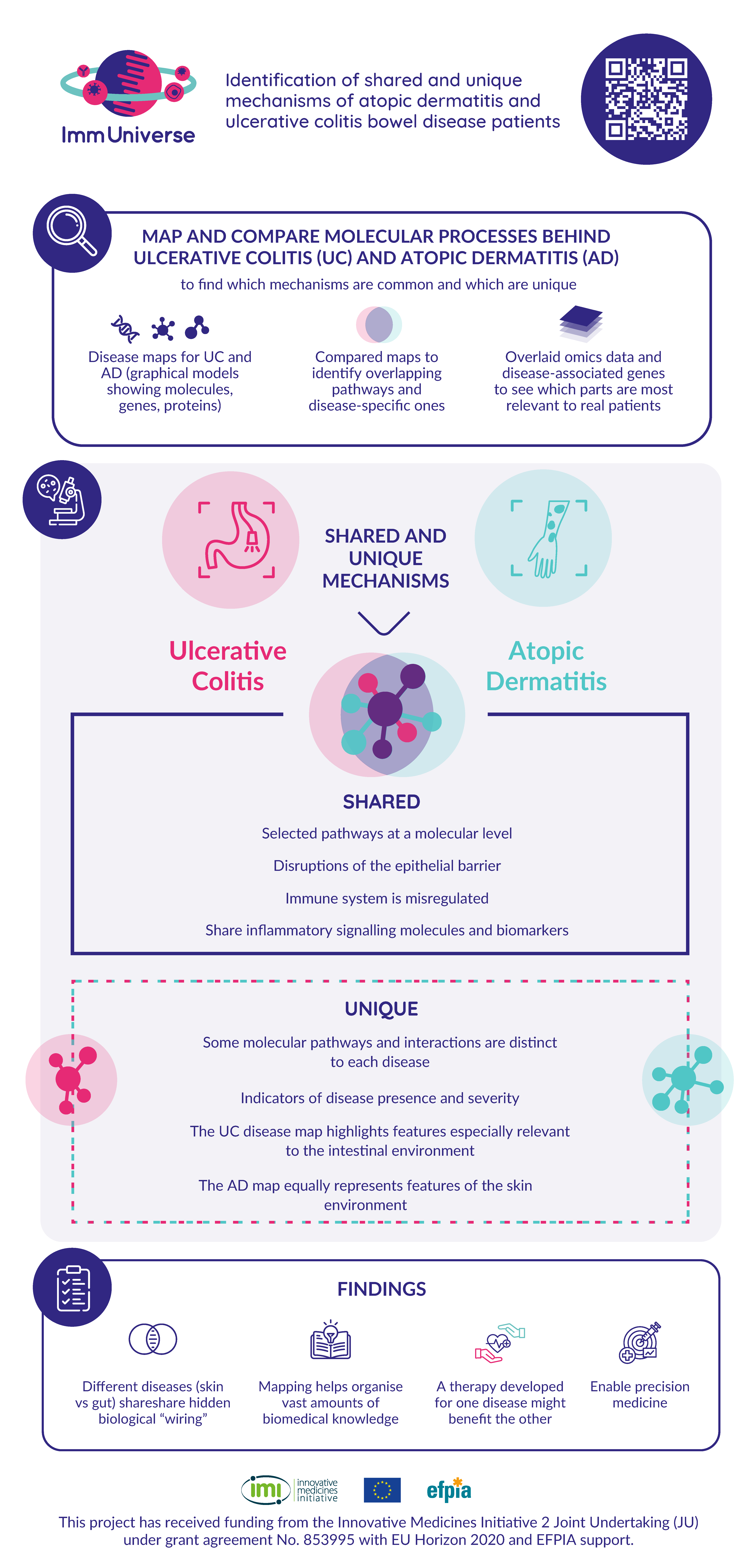
Atopic dermatitis (AD) and ulcerative colitis (UC) are two inflammatory diseases affecting different organs (skin for AD, colon for UC). They seem quite distinct, but clinical observations suggest they might share some underlying biological mechanisms. The study aimed to map and compare the molecular processes behind both diseases, to find which mechanisms are common and which are unique. This could help in understanding how the diseases develop, how they might be linked, and even suggest new therapeutic targets.
What did the researchers do?
- They built detailed “disease maps” for both UC and AD. These are graphical models showing molecules, genes, proteins and the interactions between them, based on hundreds of scientific publications.
- Then, they compared these maps computationally to identify overlapping pathways and disease‐specific ones.
- They overlaid data from gene expression studies (omics data) and disease‐associated genetic variants to see which parts of the maps are most relevant in real patients.
Key findings
1. There are shared mechanisms between AD and UC.
- Both diseases are linked to disruptions of the epithelial barrier (the layer of cells that forms a protective boundary, e.g. in skin or gut).
- The immune system is misregulated in both: including dysregulated Th2, Th1, and innate lymphoid cell (ILC) responses.
- They share inflammatory signalling molecules (cytokines) and biomarkers such as IL-13, IL-4R, IFNG, IL-18.
2. There are also unique, disease-specific components.
- Some molecular pathways and interactions are distinct to each disease, reflecting their specific tissue contexts and disease behaviour.
- The UC disease map highlights features especially relevant to the intestinal environment (barrier in colon, gut‐specific regulators).
- The AD map equally represents features of the skin environment.
3. Utility and implications of the maps.
- The maps can integrate new data and help visualise how genes or biomarkers identified in experiments fit into a larger disease network.
- Because of the overlap, the study supports the idea that therapies might be repurposed: drugs effective in one disease may also be useful in the other, depending on which pathways are shared.
- The workflow they used is reusable, meaning as new discoveries are made, they can be added into these models to refine them.
Why this matters to non-experts
- Diseases that seem different (skin vs gut) may actually share hidden biological “wiring”, understanding what’s common helps us see links between diseases.
- Mapping out disease mechanisms helps organise vast amounts of biomedical knowledge in a way that is usable for designing experiments or drugs.
- Identifying shared biomarkers or molecular players means that a therapy developed for one disease might benefit the other, shortening development time and improving chances of success.
- This work contributes to the broader goal of precision medicine, tailor treatments based on the molecular profile of disease, not just the visible organ or symptoms.
Related publication
Identification of shared and unique mechanisms of atopic dermatitis and ulcerative colitis by construction and computational analysis of disease maps
Oxana Lopata, Marcio Luis Acencio, Xinhui Wang, Ahmed Abdelmonem Hemedan, Michael J. Chao, Scott A. Jelinsky, Florian Tran, Philip Rosenstiel, Andrew Y.F. Li Yim, Reinhard Schneider, Venkata Satagopam, Marek Ostaszewski
doi: 10.1016/j.csbj.2025.09.008