Chronic inflammatory bowel diseases: Predicting the success of biologicals
Background and Context
Inflammatory bowel diseases (IBD), like Crohn's disease and ulcerative colitis, are conditions that affect 2.5 to 3 million people in Europe. Doctors treat these conditions by targeting specific chemical signals in the immune system that cause inflammation. One of these treatments uses antibodies against a substance called TNF (Tumour Necrosis Factor).
TNF acts like a signal that tells the body to start an inflammatory response, for example in the case of an injury or a disease. In the case of IBD, too much TNF can lead to excessive gut inflammation. Anti-TNF therapies are treatments that block this signal and reduce inflammation. They work by using antibodies, which are proteins that the immune system normally uses to fight off harmful substances. These antibodies are designed to recognise and bind to TNF, stopping it from signalling for more inflammation. However, not everyone responds well to this treatment, and some people experience serious side effects.
In a new study, researchers from the ImmUniverse project therefore aimed to identify biomarkers (Molecules indicating normal or abnormal processes in the body, such as genes, proteins or hormones) that can tell from the start if a patient will respond well to this therapy. Finding these markers early could greatly improve the accuracy and effectiveness of treatment plans for people dealing with inflammatory bowel diseases and improve overall quality of life.
Key Findings
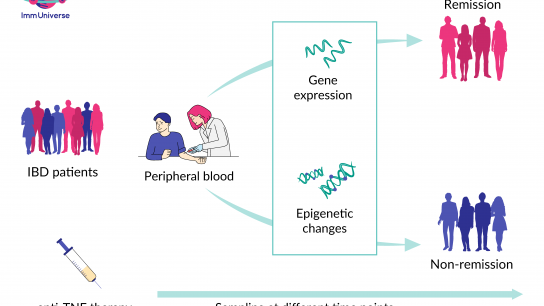
The ImmUniverse researchers examined blood samples from people with IBD before and after receiving the TNF antibody treatment. Using advanced techniques and analysis methods, the scientists looked at which genes were active in the cells and if there were any chemical changes in the genetic material.
Here is what was discovered:
- After two weeks of treatment, there were big changes in gene activity as well as epigenetic changes (Epigenetic changes are chemical and structural modifications without changing the sequence of the genetic material) in people who responded well to the treatment. About 4,000 genes acted differently in this group.
- To make sure the findings were accurate, the researchers cross-checked them with another group of people affected by IBD, which confirmed the initial results.
- Instead of testing all 4,000 genes, which would be impractical, ImmUniverse researchers identified a smaller group of about 50 genes that could reliably predict if a person will respond well to the treatment. These biomarkers were tested in a third group of people with IBD, where they accurately predicted how this group would respond to the therapy.
Real-World Benefits
The ImmUniverse research is a step toward personalised medicine for people with IBD. This means doctors can make customised treatment decisions based on each patient's unique body and molecular profile.
In the future, this could mean stopping anti-TNF therapy early for people who are not benefiting from it. This early change could save them from unnecessary side effects and speed up the start of a treatment that works better for them. Plus, it could save a lot of money within the healthcare system.
Right now, this approach is being tested in a dedicated study called GUIDE-IBD at the University Medical Center Schleswig-Holstein (UKSH), a specialised clinic in Kiel, Germany. Here, people affected by IBD are checked for specific gene activity patterns two weeks after starting the TNF antibody treatment. A team of experts reviews the results and decides if the treatment should continue. At the same time, researchers are working on making this process smoother and more practical for widespread use in regular clinics.
Related publication:
Longitudinal multi-omics analysis identifies early blood-based predictors of anti-TNF therapy response in inflammatory bowel disease
Neha Mishra; Konrad Aden; Johanna I. Blase; Nathan Baran; Dora Bordoni; Florian Tran; Claudio Conrad; Diana Avalos; Charlot Jaeckel; Michael Scherer; Signe B. Sørensen; Silja H. Overgaard; Berenice Schulte; Susanna Nikolaus; Guillaume Rey; Gilles Gasparoni; Paul A. Lyons; Joachim L. Schultze; Jörn Walter; Vibeke Andersen; SYSCID Consortium; Emmanouil T. Dermitzakis; Stefan Schreiber; and Philip Rosenstiel
doi: 10.1186/s13073-022-01112-z